Plug-Ins for Omix
Modeling & Simulation
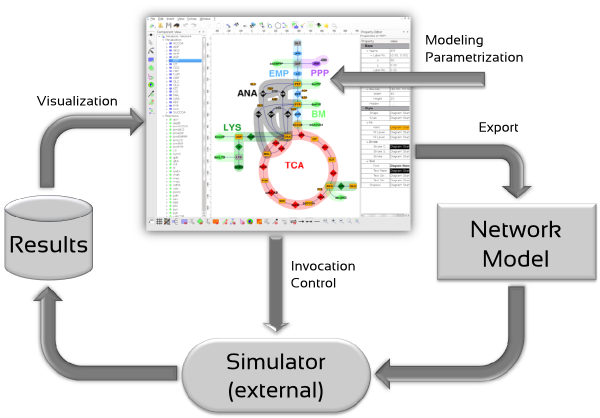
Plug-ins extend Omix to a powerful modeling tool. The network diagrams inherently contain the underlying stoichiometric model that can be used for simulation purpose. As introduced, many different plug-ins exist, that provide export functionality into various simulation formats.
Further plug-ins are available that allow to specify model parameters for different purposes and/or perform network analyses and simulation tasks directly within Omix:
-
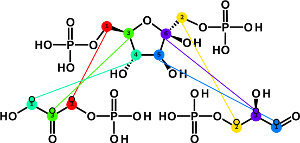 The Atomic Layer Plug-In allows to equip the reaction network
with atomic details, i.e., information about how each individual atom
is transported in the reactions. This leads to a detailed network
on atomic layer which can be used, e.g., for modeling of carbon
tracer experiments.
The Atomic Layer Plug-In allows to equip the reaction network
with atomic details, i.e., information about how each individual atom
is transported in the reactions. This leads to a detailed network
on atomic layer which can be used, e.g., for modeling of carbon
tracer experiments.
The atom mappings modeled with the Atomix Layer Plug-In are one of the main ingredients for simulations with 13CFLUX2. Other output of the plug-in can be realized on demand. - In order to produce visually identifyable and unique atom mappings, the Structural Formula Plug-In is required. This plug-in equips each metabolite with a chemical structure.
- The Network Constraints Plug-In allows to define constraint equations of the stoichiometric network. The constraints can be specified in textual manner. They serve as ingredient of 13CFLUX2 models and can be exported to other formats.
- The Free Flux Plug-In is a visual network analysis tool that helps to identify stoichiometric dependencies between reactions.
- The Elementary Flux Modes Plug-In computes and visualizes elementary flux modes in a metabolic network (see video in Example Features).
- With the Flux Balance Analysis Plug-In the correspondent network analysis can performed and directly visualized in Omix (see video in Example Features).
- The ThermoVis Plug-In allows to analyze the feasibility of reaction directions with respect to thermodynamic conditions. The thermodynamic potentials of the reactants is visualized in 3D.
Since the Omix plugin interface is public and well-documented, you can develop your own plug-ins that realize modeling features for your systems biology applications. The Omix Plug-In Development Guide explains how to develop plug-ins for Omix. The Omix API can be found here. You want to discuss your plug-in ideas or demands? Don't hesitate to contact us.
Site Overview
Plug-Ins for Omix
Imprint |
Terms Of Use |
Privacy Policy
Copyright © 2024 Omix Visualization GmbH & Co. KG. All rights reserved.
Omix® is a registered trademark of Omix Visualization GmbH & Co. KG.